24 Nov 2020
Peptide Mapping. A Beginner's Guide.
Peptide mapping is a widely used technique to examine the primary structure of biopharmaceuticals.1 It is used to help confirm the identity of a protein therapeutic and to monitor degradations such as oxidation or deamidation. Unlike intact protein analysis, peptide mapping has the advantage of being able to provide side-specific information on post-translational and chemical modifications.1 Such modifications may arise during production, processing, and storage; and since they may affect the efficacy of a biotherapeutic, they must be thoroughly characterised and controlled.
Peptide mapping usually involves enzymatic digestion of a protein. The enzyme (usually trypsin) cuts the protein up into peptide fragments at specific points in the protein sequence to produce a “fingerprint” of fragments, which are then chromatographically separated and identified by MS detection.2 Figure 1 shows an example of the peptide fragments that are created when Herceptin (trastuzumab) is digested with the enzyme Trypsin, which cleaves following a lysine or arginine amino acids in the protein.3 It is important that as much of the protein sequence is covered as possible by the peptide map, since this enables confident protein identification and can provide additional information beyond that obtained at the intact protein level.2
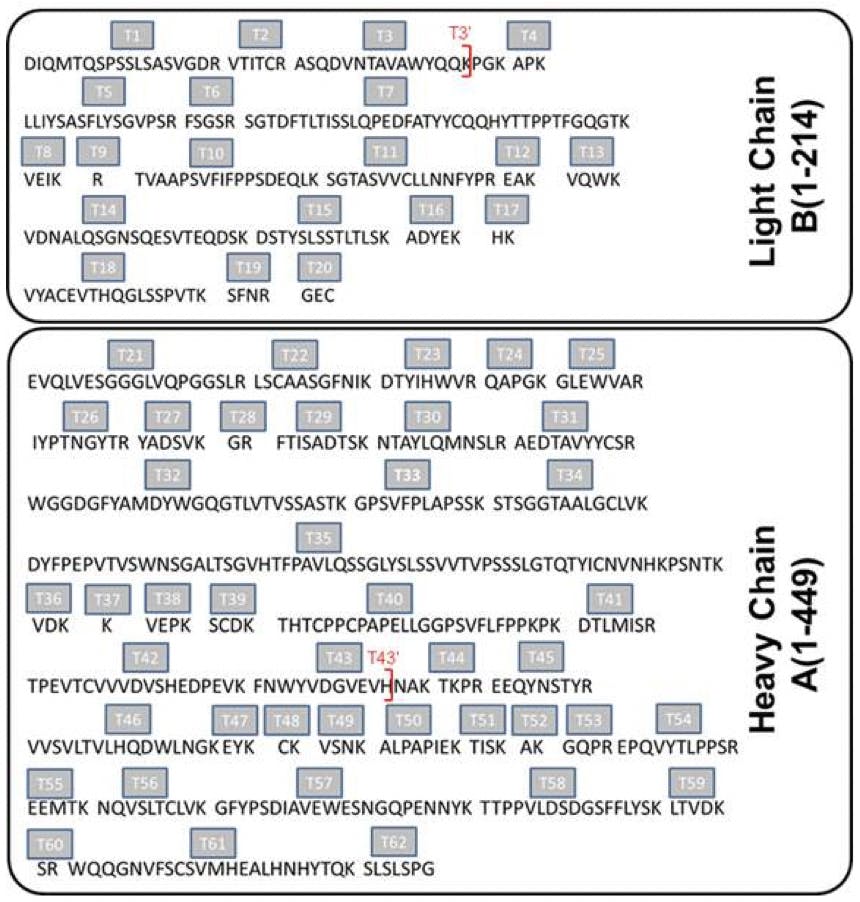
Figure 1 illustrates the 62 resulting Herceptin (trastuzumab) peptides following trypsin digestion; 20 from the light chains (Lc) and 42 from the heavy chains (Hc).3
The unrivalled resolving power of reversed phase chromatography (RPC) has made it the predominant HPLC technique for separating out peptide fragments.2 Although it is the technique we will focus on here it is worth remembering that, depending on the protein being studied and the experimental objectives, other techniques may also be necessary. One example of this is when working with small polar peptides that are not well retained by RPC. In this case, hydrophilic interaction liquid chromatography (HILIC) may be needed to get sufficient separation to allow identification of the peptide fragments. A typical peptide mapping profile may include over 100 peaks that each represent an individual peptide and their derivatives. Knowledge of sample preparation methods along with efficient separation methods and protocols is required when developing a new peptide mapping strategy 2
The objective of this article is to discuss some of the areas that are important for generating peptide maps by RPC, and highlight considerations for optimising separations to achieve the best possible sequence coverage.
Sample preparation
The pre-treatment required for your sample will depend on the size and confirmation of your protein2. It might be necessary to enrich the sample or to separate the protein from added substances used in the formulation of the product, since these may interfere with either the actual protein digestion or the subsequent LC or LC/MS. Each protein will have its own clean-up protocol, but dialysis and desalting are typically required for most protein samples to ensure they are compatible and optimized for digestion. Since peptide mapping is routinely done by LC/MS, desalting is necessary to reduce salts (especially sodium and phosphate) to avoid interference with detection. Dialysis is an established procedure for reducing the salt concentration in samples and can also be used for buffer exchange and small molecule removal. It requires filling a dialysis bag (or sometimes a cassette), which is a membrane casing of a defined porosity, with your sample. The bag is placed in a bag of water or buffer where the concentration of salt will equilibrate through diffusion. Porosity is chosen so that large molecules, such as the protein of interest, cannot diffuse through the membrane and remain in the bag. Once the equilibration process is complete the sample can be retrieved from the bag. Although effective, this is a slow process and, since various factors can affect the equilibration process, the final salt concentration can be somewhat variable. A more robust and well-defined method for desalting samples prior to digestion is Gel Filtration (GF). This is a non-adsorptive chromatography technique that separates molecules based on molecular size. Like dialysis, this can be used for desalting or for buffer exchange, where the sample buffer is replaced with a new buffer.
Protein digestion
To first stage in peptide mapping is to break the protein down into peptide fragments by proteolytic enzymes. A good understanding of this process will help to ensure the maximum sequence coverage and sensitivity are achieved. Often this step requires its own development to provide a stable sample for LC injection; however, several common approaches are followed.2 The key steps of protein digestion are summarised in Table 1.
| Procedure | Intended Effect | General method |
| 1. Selection of Cleavage Agent | Specific cleavage requirement | None |
| 2. Reduction and Alkylation | Reduction of disulphide bonds Alkylation caps SH groups to prevent reformation of disulphide bonds |
Reduction DTT, 45 min, 60°C Alkylation IAM, 1hr in the dark |
| 3. Digestion Process | Cleavage of proteins | Digestion pH 8, 37°C, overnight Quenching: addition of TFA |
Table 1. Common steps in protein digestion.
Although there are chemical methods that can be used for the cleavage of peptide bonds, enzymatic approaches are more commonly employed for peptide mapping.2 This is due to their site-specific cleavage locations, which produce very predictable peptide fragments. Trypsin is the most common agent used for peptide mapping due to its well-defined specificity. Trypsin hydrolyses only peptide bonds in which a carbonyl group is followed either by an arginine (Arg) or a lysine (Lys).2 This cleavage pattern can be observed in Figure 1. Depending on the protein amino acid sequence, other enzymes may also need to be used to improve sequence coverage of the resulting peptide mapping. Some of the most frequently used enzymes are summarised in Table 2 along with their cleavage site.
| Enzyme | Site of Cleavage |
| Trypsin | Lys, Arg (C) |
| Chymotrypsin | Phe, Trp, Tyr (C) |
| Asp-N-protease | Asp, Glu (C) |
| Pepsin | Leu, Phe, Trp, Tyr (N) |
| Elastase | Ala, Gly, Ser (C) |
| Endoproteinase Lys C | Lys (C) |
Table 2: Common proteolytic digestion enzymes and their specific cleavage sites.
The importance of selecting an appropriate protease enzyme can be see in Figure 2 below. The comparative peptide maps for Myozyme digested with either LysC or trypsin are shown.3 The peptide map produced by LysC not only has fewer peptides (16 compared with 58 in the trypsin map), but they are also much larger in size. The increase in size and therefore in hydrophobicity results in the peptides eluting later in the chromatogram under identical reverse phase conditions.3
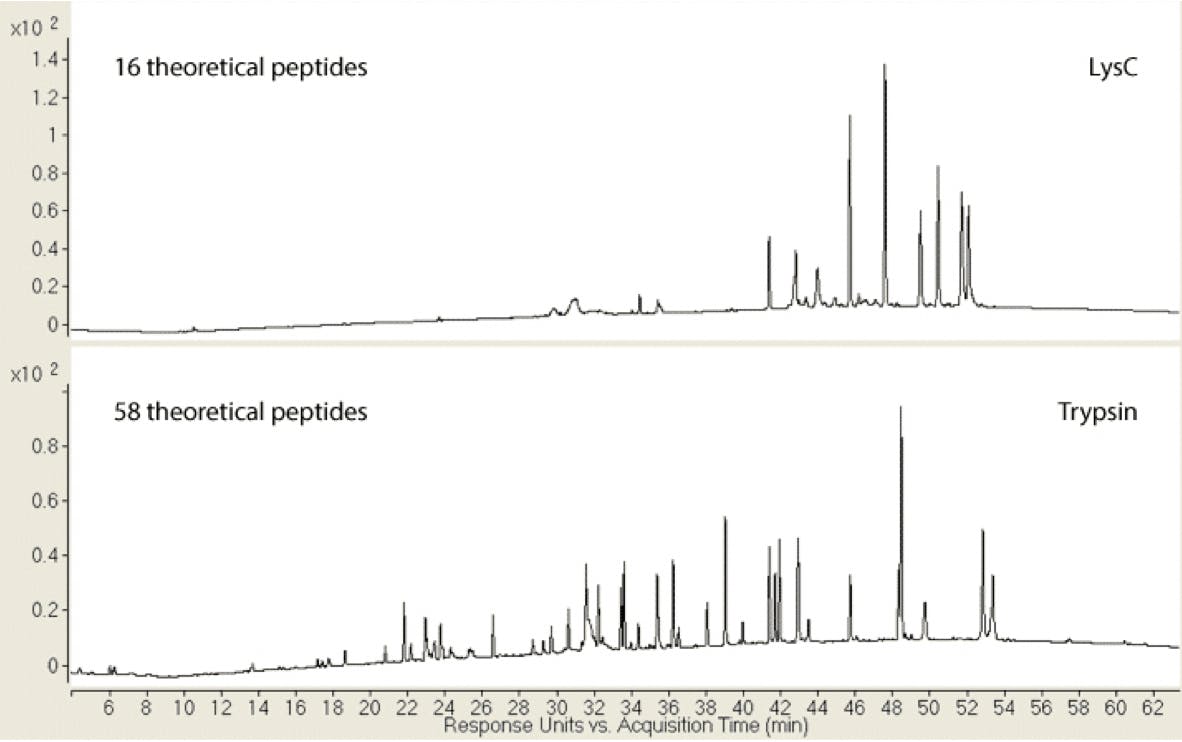
Figure 2: Myozyme peptide maps with either LysC (top chromatogram)) or trypsin proteolysis (bottom chromatogram).3
Reduction and Alkylation
The first stage in the reduction step is to denature the protein to remove the higher order structure and expose the many internal disulphide bonds.3 Denaturation is often achieved by high temperature or chemical reaction.2 Once the disulphide bonds are exposed they can be reduced using a reagent such as 1,4-dithiothreitol (DTT), mercaptoethanol, or tris(2-carboxethyl)phosphine (TCEP).3 Once the disulphide bonds are exposed they can be reduced using a reagent such as 1,4-dithiothreitol (DTT), mercaptoethanol, or tris(2-carboxethyl)phosphine (TCEP).3 Most commonly denaturation and reduction are combined (i.e. heating the sample in the presence of DTT) which avoids the problem of renaturation during the process. Following this step, alkylation of the cysteine is needed to avoid reformation of the disulphide bonds in the protein (renaturation). To this effect, the protein is incubated with an alkylating agent such as 2-iodoacetamide (IAA).3
Digestion Process
Commonly, digestion is performed at an optimal pH and temperature for the proteolytic enzyme being used, typically pH 7.5 - 8.5 at 37 °C. It is worth highlighting that when model proteins are digested in a mixture vs separately, less effective digestion has been observed. This may be due to increased competition for enzyme cleavage sites when more different types of proteins are digested together. Several parameters need to be optimised to ensure the completeness and effectiveness of the digestion of a protein sample, for example it is crucial to optimise reaction time, pH, temperature, and concentration of cleavage agent used.2
Peptide digest clean-up
Following protein digestion some clean-up and/or enrichment is typically required before analysis. The method used will depend on the specific sample type and objective of the analysis. Enrichment will need be targeted towards the specific analysis being performed. Examples include enrichment for specific post-translational modifications (PTMs) by affinity purification using specific antibodies or ligands designed to bind to peptides with PTMs such as phosphorylation, ubiquitination, and glycosylation.2 Salts, buffers and detergents from the digestion and enrichment process will generally need removing before analysis is performed. Desalting is usually done using either graphite or C18 tips or columns, whereas detergent can be removed via precipitation or affinity columns.2 If required a concentrator of the appropriate molecular weight cutoff (MWCO) can be then be used to concentrate dilute samples prior to HPLC analysis.2
Peptide Separation
Due to excellent resolving power, speed and use of volatile mobile phases (compatible with mass spectrometry) RPC is usually the first-choice technique for peptide separations. Column selection, quality and mobile phase selection and detection requirements are all important to consider during method development to ensure you a achieve a reproducible and robust RPC method for peptide mapping. Although a full discussion around peptide mapping method development options is beyond the scope of this article, column selection is a crucial choice worth mentioning. Pore size, particle type and size, and the bonded phase chemistry and stability all play important roles in achieving a well resolved peptide map. At Element we have a wealth of technical expertise in this area so we can support you with these method development decisions. Email our technical team for more details.
Since the peptide fragments generated by protease digestion are usually quite short, the preferred column pore size for a peptide mapping column ranges from 100-160 Å. Smaller pore sizes may be useful if dealing with particularly small peptide fragments, but have limited usefulness when trying to achieve retention across a broad spectrum of peptide hydrophobicity. Given the complexity of the separation, a high efficiency column is required. This can either be achieved by choosing a sub-2µm particle, or a superficially porous particle (SSP). SSP columns have become increasingly popular for peptide mapping since they address the limitations of peptide diffusion. SSP columns allow the separation of larger molecules at high linear velocities without the high system back pressure associated with smaller particles. Figure 3 shows an example of a high-resolution peptide map achieved using a SPP column.
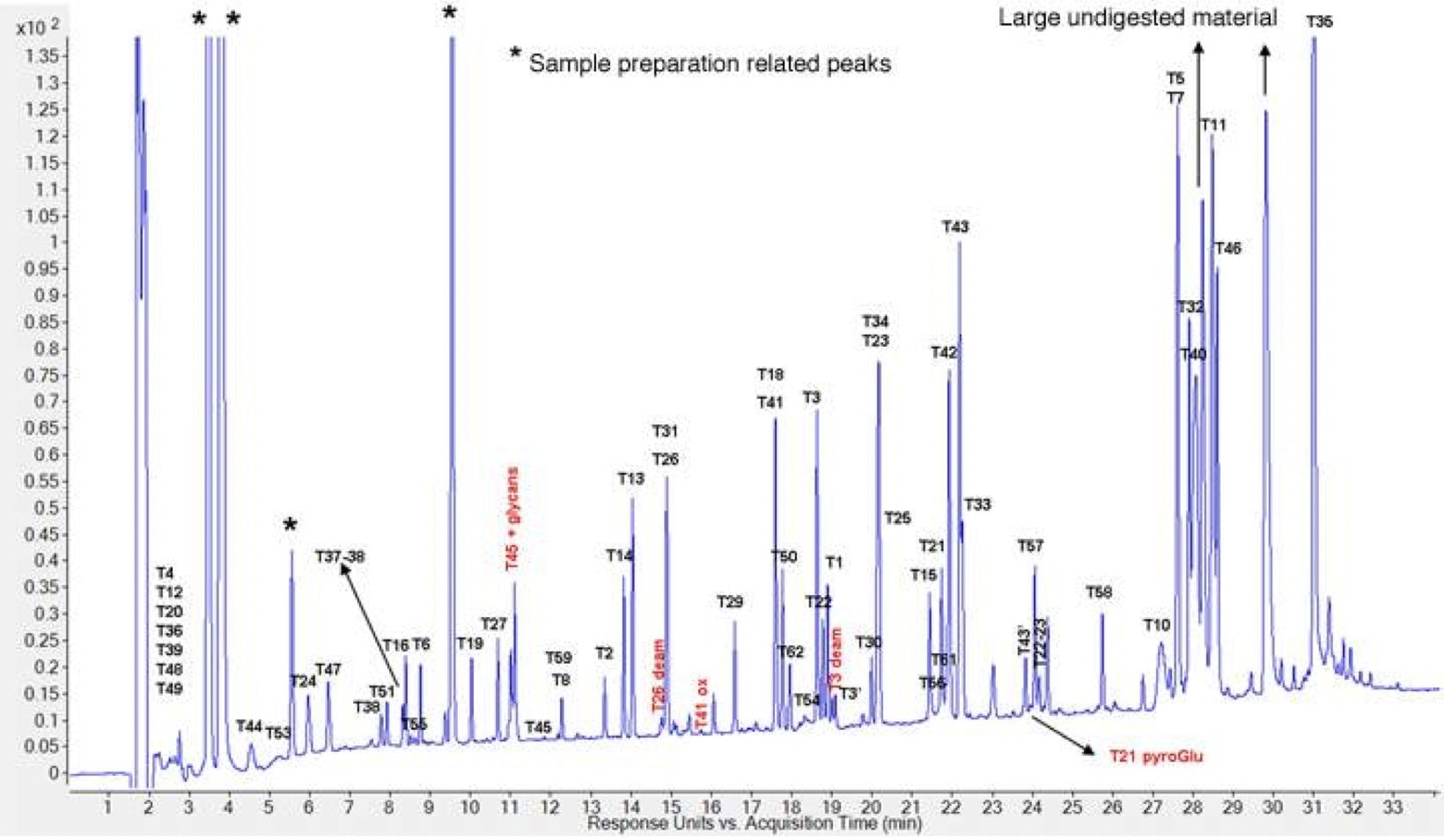
Figure 3: Typical Herceptin peptide map. Agilent AdvanceBio Peptide Mapping 2.1mm x 250 mm x 2.7 µm column, mobile phase A: 0.05% TFA, mobile phase B: 0.05% TFA in acetonitrile, flow rate 0.3mL/min, UV 214 nm, gradient 1-45 %B 2-35 minutes, 60 oC.3
Although a C18 reverse phase column is the most common chemistry used for peptide mapping, alternative phases can provide useful selectivity differences that can be useful for resolving overlapping peaks and improving peptide mapping coverage as demonstrated in Figure 4 below.4
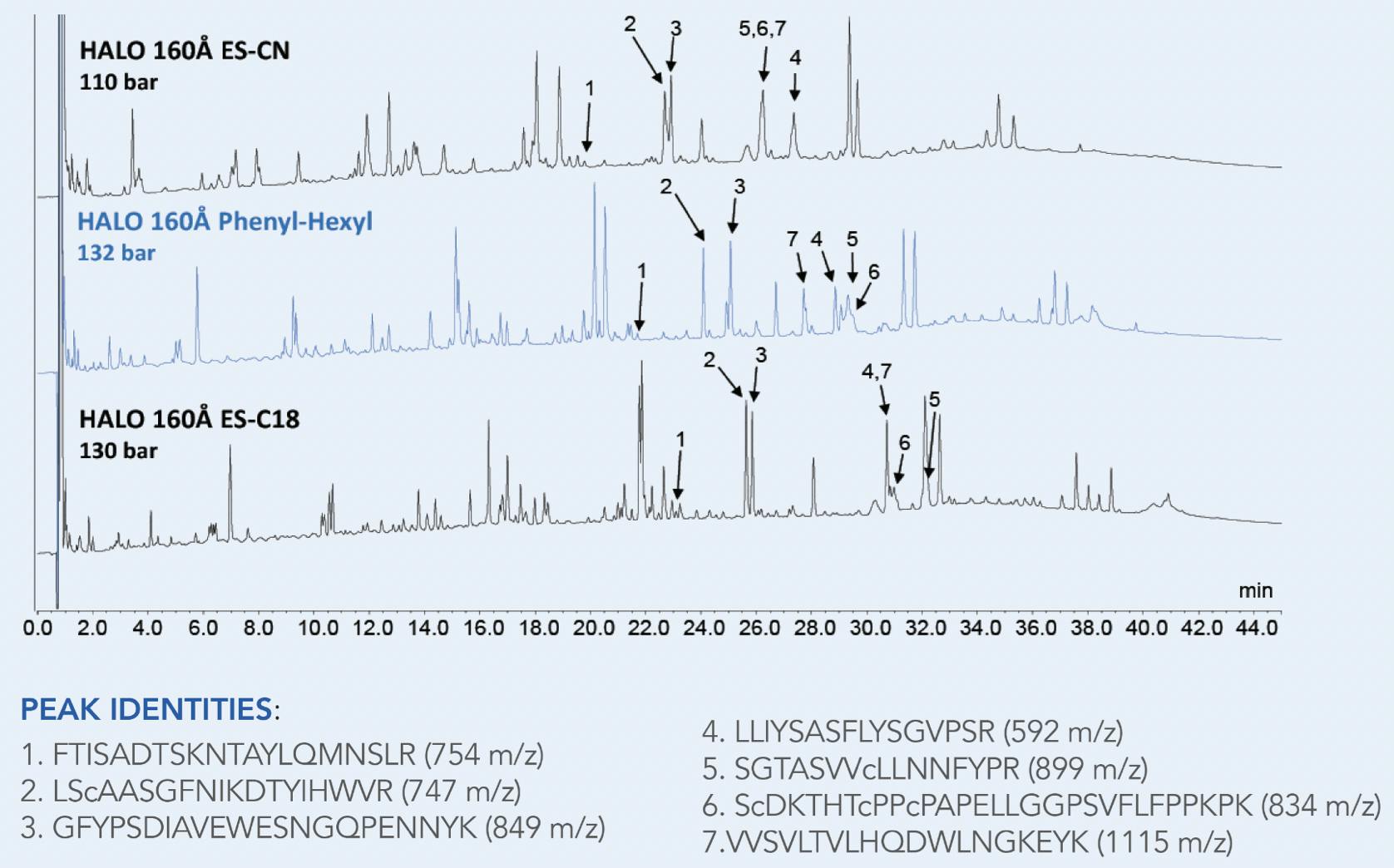
Figure 4 Enhanced separation of peptide fragments from a tryptic digest achieved by using alternative phase chemistry. In this case a Phenyl-Hexyl column provides improved resolution between fragments 2 and 3 compared to a ES-CN or an ES-C18 column.4 (Column dimensions: 2.1mm x 100 mm x 2.7 µm, mobile phase A: water + 10 mM difluoracetic acid (DFA) mobile phase B: ACN + 10 mM difluoracetic acid, flow rate 0.3 ml/min, UV 220 nm, gradient: 2-50 % B in 60 min at 60 °C)
Detection by UV and Mass Spectrometry
UV detection for peptides is usually carried out at 210-220 nm and or/ 280 nm (Figure 5). Tryptophan, tyrosine and phenylalanine amino acids are sensitive at 280 nm whereas detection at 210 nm is relatively unselective. Although detection at 210 nm will also detect other biological molecules in the sample matrix, it is 2-4 fold more sensitive than 280 nm – so detection at 210 and 280 nm is often done in parallel. Figure 5 shows a peptide mapping separation comparison between detection at 220 and 280 nm, demonstrating the differences in absorbance sensitivity and UV peak profiles.
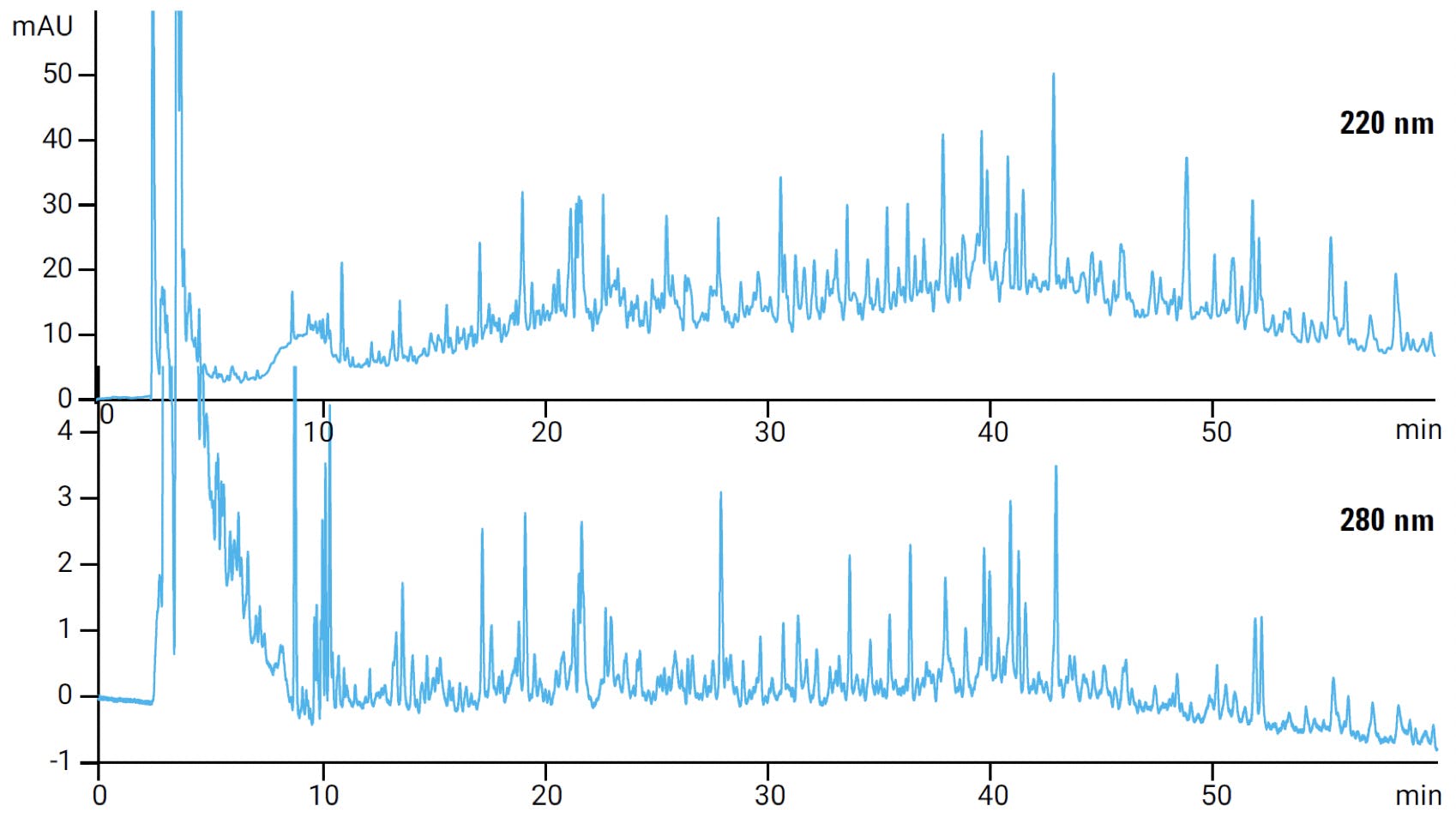
Figure 5 Peptide mapping profile of a digest at 220 nm (top) and 280 nm (bottom) on an Agilent 1290 Infinity LC.
Mass spectrometry has become an important method of detection for characterizing peptides and peptide maps, making it possible to quickly determine the identity of peptide fragments. This is particularly important in the biopharmaceutical industry where establishing and monitoring the sequence identity of a therapeutic target is critical.3 Peptides can be analysed through direct infusion of the isolated peptides, or by the use of online LC/MS. The results can be correlated to the protein, to confirm the specific amino acid sequences covered by the peptide map and the protein identity. By comparing the measured masses from the MS information to the predicted values from the intact protein or protein database, the mass and sequence coverage information can be used to identify the protein.3 The goal of characterization through peptide mapping is to achieve at least 95 % sequence coverage of the theoretical composition of the protein structure.
Summary
Some of the key points in peptide mapping sample preparation and separation have been introduced in this article. At each point in the process, optimisation and consideration is needed to ensure that a suitable method is customised to your protein and sample matrix. Although standard methods have been developed for peptide mapping, the unique composition of each protein and sample means that even a generic method will need some optimisation to suit your sample and gain the maximum information from this technique. At Crawford we can supply a range of consumables for peptide mapping as well as advice and support with method development and optimisation through our in-house technical team. Struggling with where to start your method development or having issues with sample preparation? Get in touch with our technical team who can provide advice and support on the best products and protocols for your particular method.
References
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5830484/
- https://www.agilent.com/cs/library/applications/compendium-peptide-advancebio-5994-0037EN-us-agilent.pdf
- https://www.chromacademy.com/channels/bio-chromatography/technique/reversed-phase-introduction-and-peptide-level-analysis/
- https://advanced-materials-tech.dcatalog.com/v/Halo-Bioclass-Catalog---Searchable/?page=14