27 Jan 2020
The Hofmeister Series
The influence of salt concentration on the solubility of proteins, nucleic acids, and other macromolecules has been known since at least the early nineteenth century. Before the introduction of chromatographic techniques, “salting out” was the most common method of protein purification.
“Salting out” is described as the phenomenon where the solubility in water of a substance decreases with increasing salt concentration. “Salting in” is used when the solubility of a substance increases with increasing salt concentration. Setschenov published an important equation in 1892, while investigating the effect of salts on the solubility of CO2:
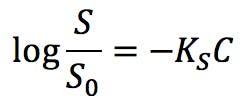
Where S0 is the solubility of a substance in pure water, C is the concentration of salt, and KS is the salting out coefficient, or Setschenov constant. Substances with positive KS experience salting out, whereas substances with negative values are susceptible to salting in.
Due to their complex structures, proteins have a more complex solubility dependence on salts than simple organic molecules. At very low salt concentrations, proteins are poorly soluble in water and tend to agglomerate. Increasing salinity improves solubility, and proteins are “salted in” according to Setschenov’s law. It is assumed that agglomeration takes place because charged amino acid residues on the surface of the proteins are poorly shielded in pure water, resulting in strong intermolecular attractions that eventually cause the aggregates to precipitate. With increasing concentration of ions in the medium, electrostatic attractions are shielded and the protein becomes more soluble.
However, the solubility of the protein stabilises when reaching a certain salt concentration, and eventually decreases at higher salinity: the proteins “salt out” at high salt concentrations.
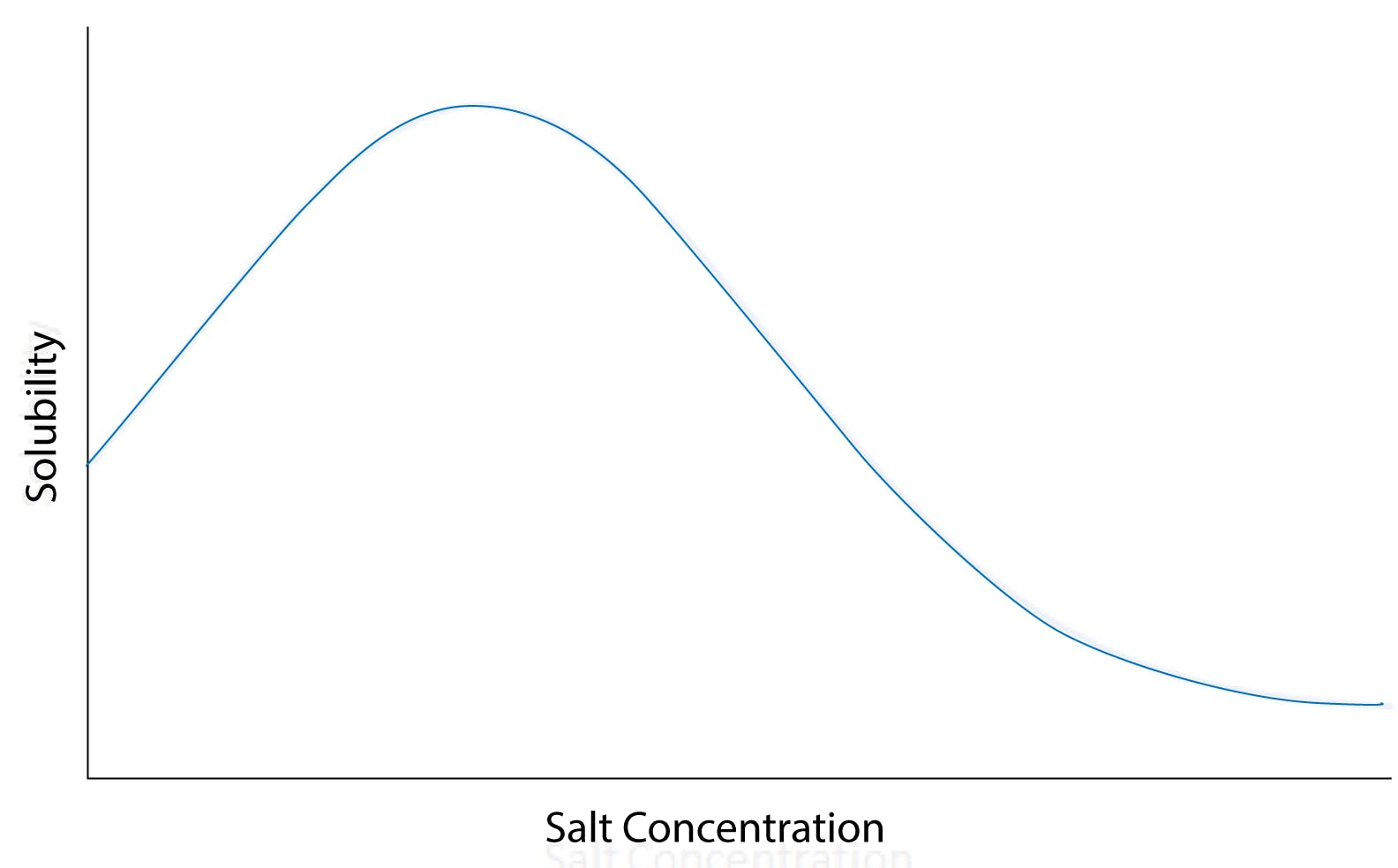
Fig 1: Solubility versus salt concentration curve for a typical globular protein, illustrating salting-in and salting-out at low and high salt concentrations, respectively.
Before the widespread use of chromatography, salting out was the preferred method for protein purification: a protein solution would be forced to precipitate by adding a sufficient amount of salt, and the protein could then be recovered from the supernatant by careful filtration.
Hofmeister developed his influential series of ions in 1888, while measuring the salt concentration necessary to precipitate proteins from egg white. As he experimented with different salts, he observed that some were more efficient at salting out (i.e. they could cause proteins to precipitate at lower concentrations) than others. He empirically found that common anions could be ordered from highest to lowest precipitating capacity:
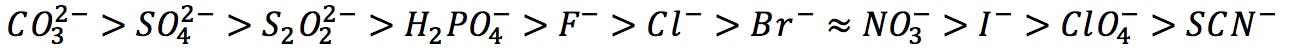
Hofmeister series of anions
Similarly, he ranked common cations from highest to lowest protein precipitation capacity:
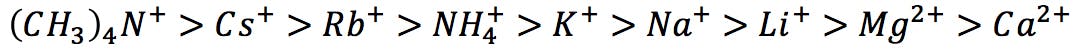
Hofmeister series of cations
The differential effect of different electrolytes on solubility does not only affect proteins. Small molecules, especially lipophilic neutral species, are strongly affected as well. The following graphs, for example, show the salting out effect of several anions and cations on aqueous solutions of benzene:
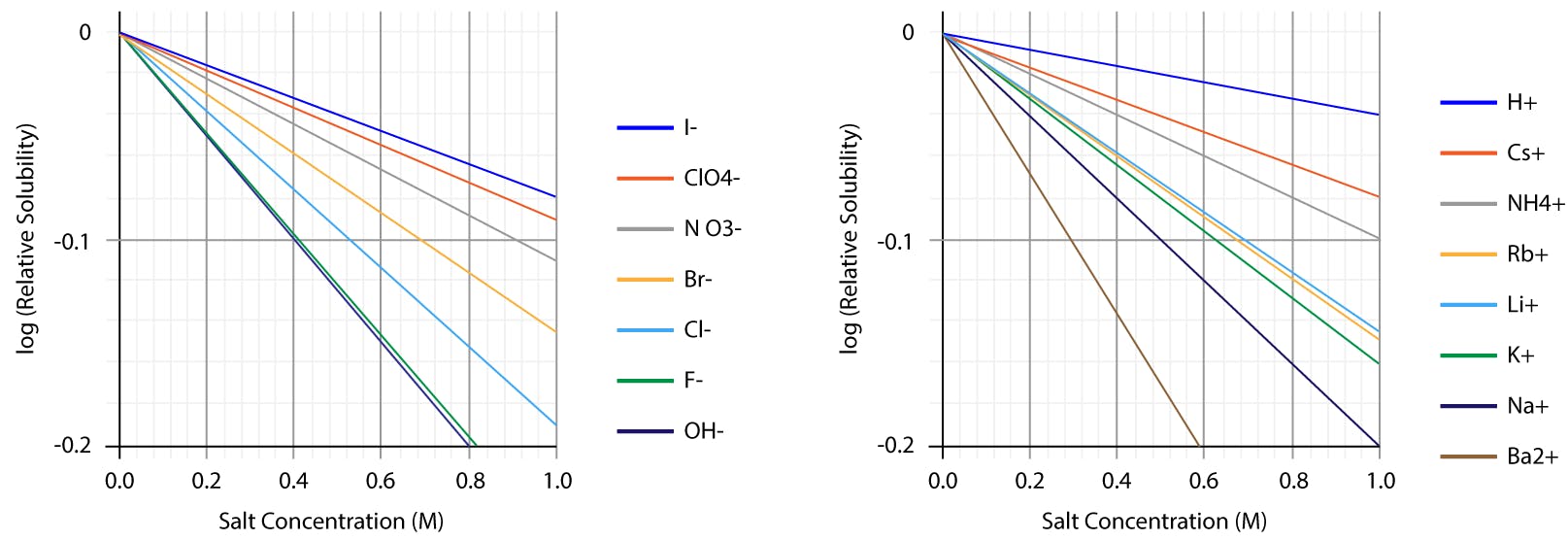
Fig 2: Salting out curves of benzene aqueous solutions, as observed for different anions and cations. Note the linear trends, consistent with Setschenov law.
Although the precise effect is solute-specific, there is a certain correlation between these electrolytes on benzene and the series that Hofmeister found for proteins. The Hofmeister series is of interest in a wide range of disciplines, including biochemistry, microbiology, physiology, aquatic and atmospheric chemistry, food science and technology, polymer chemistry, etc.
Several theories have been put forward over the last century to explain the salting in and salting out effects of ions in solution, and the true mechanisms is still badly understood.
Setschenov interpreted the effect as weak interactions between the molecules of solute and its chemical environment, where salt ions and water molecules underwent a constant exchange on the surface of the molecules. Salting out of simple substances like benzene is well explained by this model, and the empirical solubility curves fit well the Setschenov equation. However, as we have seen proteins do not obey Setschenov’s law at high salt concentrations, demonstrating that other mechanisms must be at play.
An alternative theory, the site binding model, considers the salt ions as chemical reactants which bind to specific sites of the solute molecule. This model is suitable for molecules with ionisable and polar functional groups, where electrostatic ion-ion attraction and ion-dipole forces can explain the binding mechanism. It is not applicable, however, to nonpolar molecules, such as alkanes.
A third model, approximately contemporary of Hofmeister’s work, considered that ions in solution affected solubility by organizing or breaking the structure of bulk of the water solvent. This theory is known as the cavity model.
It is well known that water molecules can form ordered structures under certain thermodynamic conditions. These structures arise from the directional nature of hydrogen bonds, and descriptions of these so-called clathrates go as far back as Humphrey Davy. The structure shown below is made up of 28 water molecules. Each vertex indicates an oxygen atom, and the covalently bonded hydrogen atoms would be positioned along the edges. A hydrogen bond between each hydrogen and the oxygen of an adjacent molecule would account for the ordered structure. The twelve pentagons and four hexagons define a cavity around 6Å in diameter, large enough to accommodate a short hydrocarbon molecule.
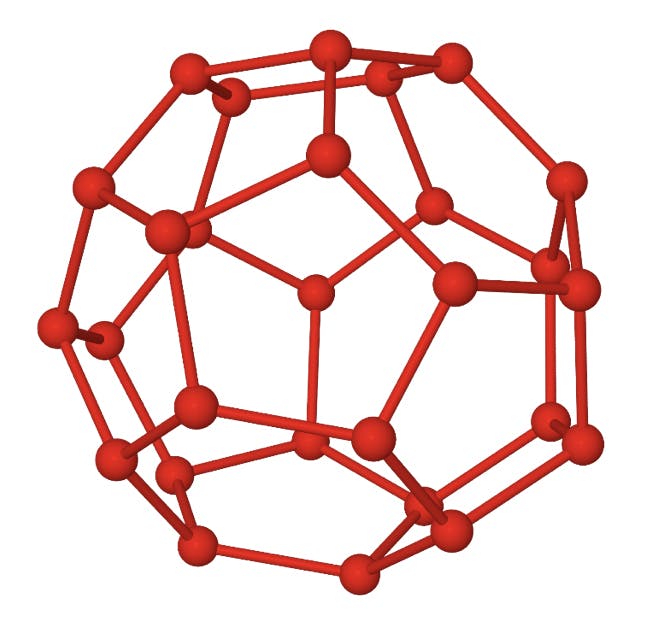
Fig 3: Structure of a 51264 water clathrate.
The cavity model is closely related to the hydrophobic effect: whilst water molecules can interact directly with polar and ionic functional groups on the surface of a protein and facilitate its solubility, the hydrophobic residues of non-polar amino acids would repel water molecules and have the opposite effect. The figure below illustrates this principle for lysozyme, one of the proteins that Hofmeister would (unknowingly) have purified from egg:
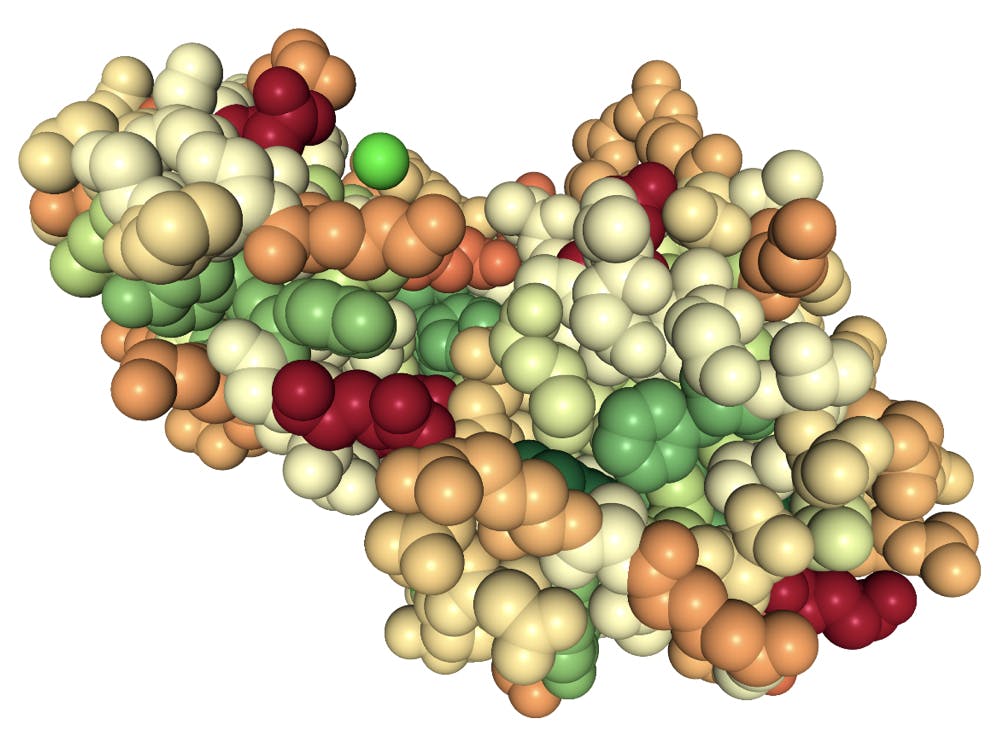
Fig 4: Space fill model of a lysozyme protein. Colours indicate polarity of the amino acid side chains: red highly polar, green highly hydrophobic.
The groups in dark red are highly polar glutamic acid residues; the groups in green are hydrophobic residues phenylalanine and tyrosine. The bright green sphere represents a chloride ion coordinated by glycine and lysine (pale orange) residues. Note that most of the hydrophobic groups are folded inside the protein, but also that a large fraction of the surface is not particularly polar.
According to the cavity model, the hydrophobic regions of the protein would be encased in clathrate-like structures. These highly ordered structures are entropically unfavourable, and in pure water the protein tends to aggregate to minimise the hydrophobic area exposed. If enough aggregation occurs, the protein flocculates out of solution.
The anions on the right of the Hofmeister series (perchlorate, thiocyanate, iodide) are among the most effective salting-in agents. These anions are large and have a low charge density. They are capable of binding solutes through nonlocalized dispersion forces, and thus add charges to the surface of the protein. Large cations with delocalised charges include pyridinium, guanidinium, or tetraphenylphosphonium, which can establish similar dispersion bonds with proteins. In that sense, these species behave like surfactants, enhance the solubility of the molecule, and have a strong salt-in effect.
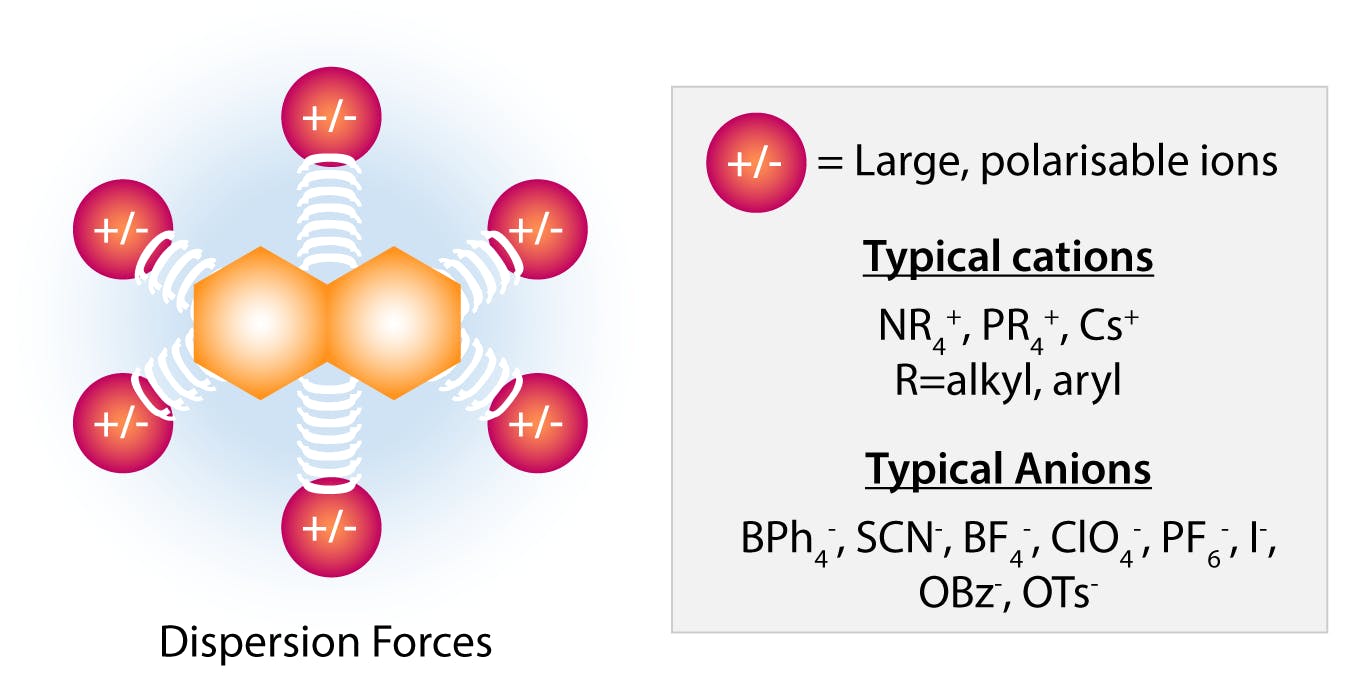
Fig 5: Salting in effect by large “soft” anions and cations. (Adapted from Hyde et al, 2017)
Small cations with highly charge densities, such as those on the right of the Hofmeister series, can also salt in proteins, but in this case the binding mechanism is specific ion-dipole forces with polar residues on the molecular surface, as proposed by the site binding model.
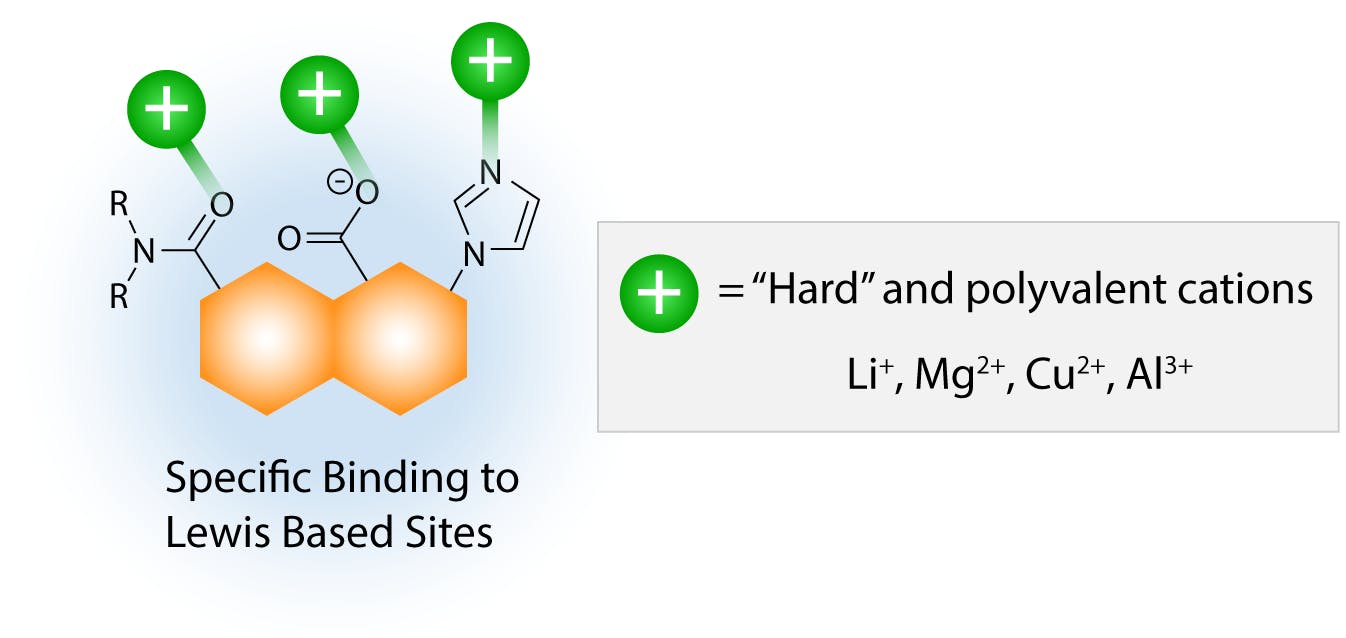
Fig 6: Salting in effect by small “hard” cations. (Adapted from Hyde et al, 2017)
Because of the specific nature of these interactions, the relative salting-in effect of different cations depends on the functional groups present on the solute or protein, and it is more difficult generalise the salting sequence. The salting strength of these cations is also in general weaker than that of large, polarisable ions, possibly due to the fact that specific interactions are stoichiometric, whereas dispersion interactions can involve various ions simultaneously. With increasing concentration of ions in solution, the salting-in effect eventually reaches a tipping point when the high ionic strength of the environment is incompatible with the lipophilic regions on the protein surface. As the hydrophobic effect becomes stronger, the protein tends towards aggregation again to minimise the area of interface with the ionic environment.
An alternative interpretation is that high salt concentrations polarise water molecules to solvate the ions, strengthening the ordered structure of the clathrate. As more water is excluded from the molecular interface, the proteins are isolated from the ionic medium and Brownian collisions result in rapid aggregation. Eventually, the aggregates precipitate out of solution.
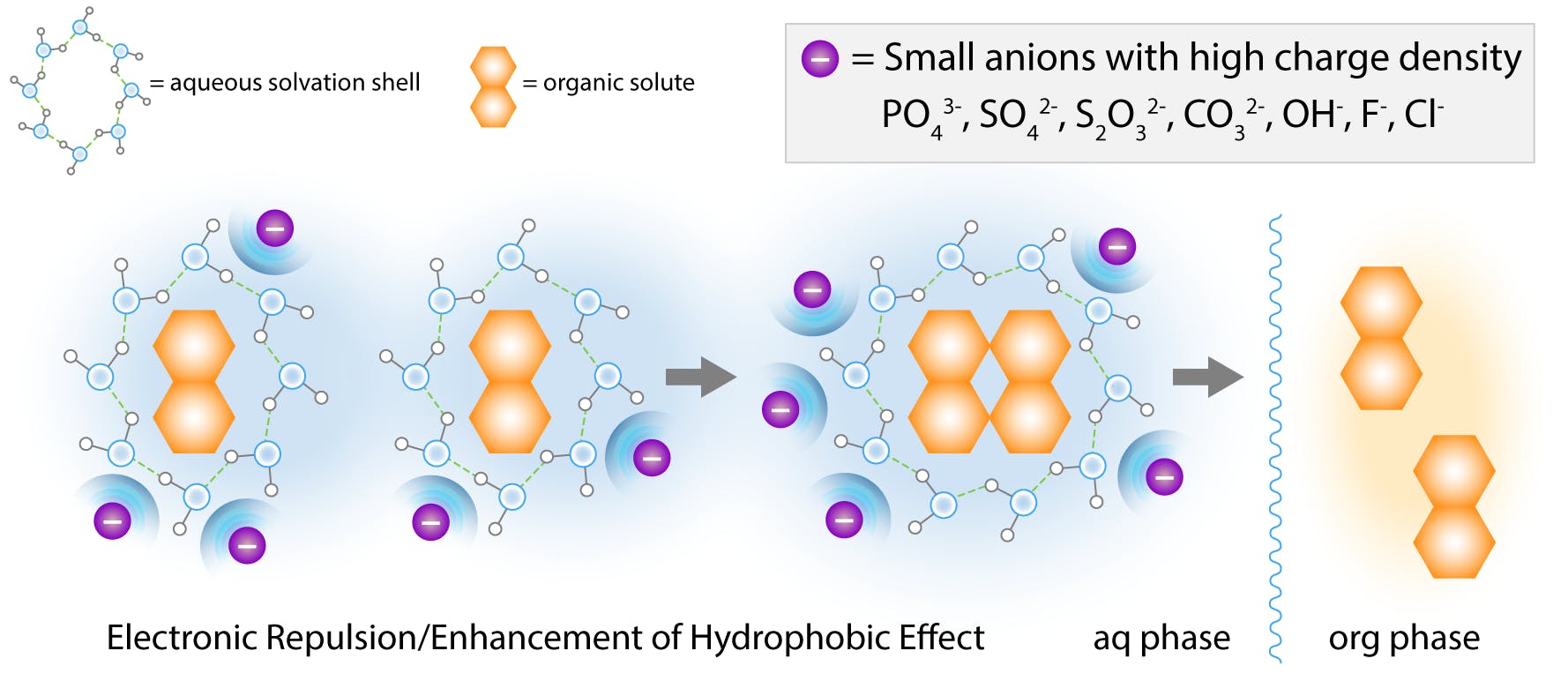
Fig 7: Salting out effect by small “hard” anions according to the cavity model. (Adapted from Hyde et al, 2017)
Small “hard” ions (high charge density) on the left of the Hofmeister series exhibit the strongest salting-out effect. These ions attract a large amount of solvation water, and are highly hydrated. They are also known to increase the surface tension of water.
These properties enhance the hydrophobic effect and the formation of cavities, and these ions have traditionally been called kosmotropes, “makers of order”.
Large “soft” ions, on the other hand, reduce water surface tension and disrupt solvent cavities, for which they are called chaotropes, “makers of chaos”.
Although the cavity model has been taught in chemistry schools for decades, recent studies suggest that it is only one of many contributors to protein stability, and that the picture is somewhat more complicated. It is now believed that strong chaotropes such as thiocyanate and iodide bind preferentially (salt in) the polar peptide groups, promoting the unfolding of the peptide chain to expose the complete backbone of the protein. In doing so, they denature the protein and increase its solubility.
This theory would explain why guanidinium thiocyanate is a strong denaturant, whereas guanidinium sulphate is not. Kosmotropes, on the other hand, would stabilise the protein structure by salting out non-polar groups through the cavity model and possibly through weak non-specific interactions, ion-dipole and electrostatic interactions with polar residues, etc. The emerging picture is that of a complex equilibrium of competing mechanisms that depend on specific groups within the peptide chain, as well as the nature of the ions and ionic strength of the solution.
Even though the specific mechanisms involved in salting in and salting out are not fully understood, the following empirical guidelines are well established:
- Small, multiply charged “hard” anions have a strong salt-out effect.
- Small, “hard” cations may or may not have an effect depending on solute functional groups .
- Large anions and cations with a diffuse charge density (lipophilic character) always have a salt in effect.
- Combinations of anions and cations exhibit additive effects. A highly lipophilic anion or cation is likely to dominate and have a prevalent salt-in effect.